Note
Go to the end to download the full example code
Classifier comparison
A comparison of a several classifiers in imbens.ensemble
on synthetic datasets. The point of this example is to illustrate the nature
of decision boundaries of different imbalanced ensmeble classifiers.
This should be taken with a grain of salt, as the intuition conveyed by these
examples does not necessarily carry over to real datasets.
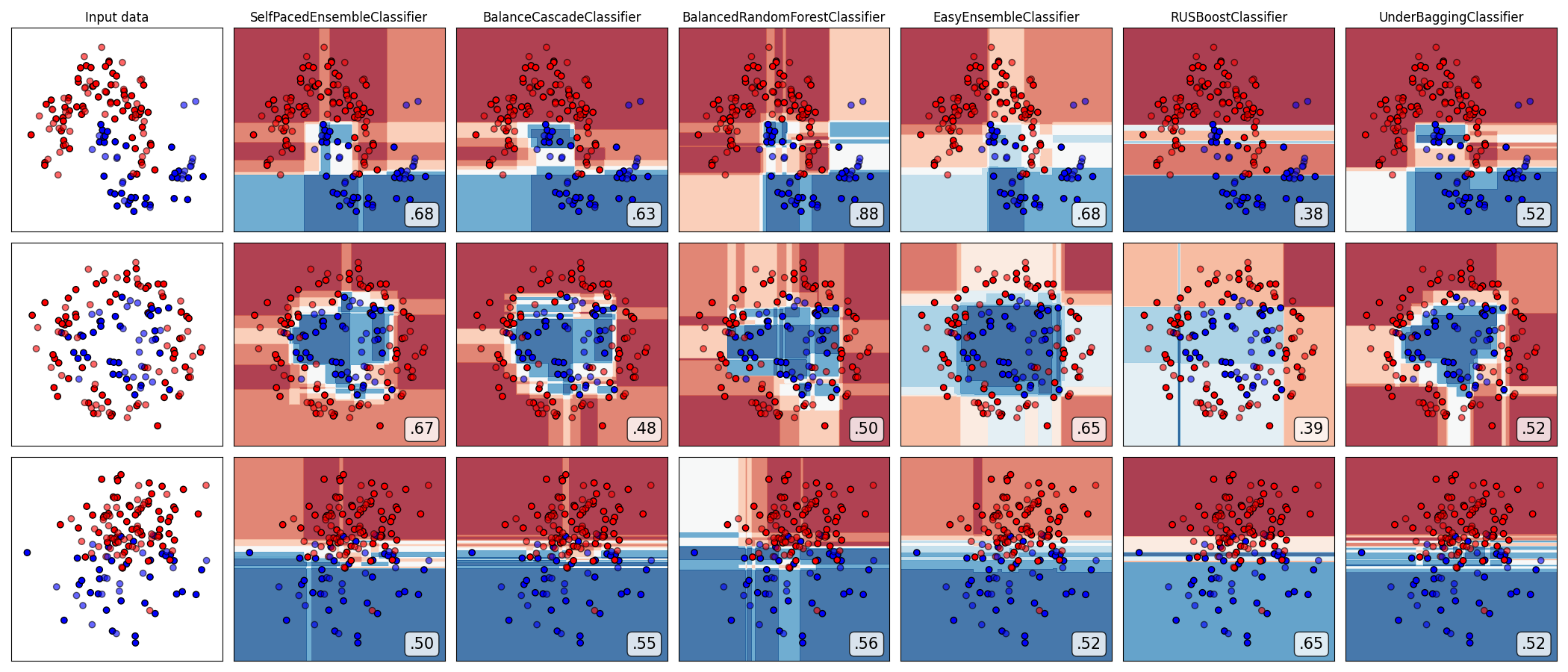
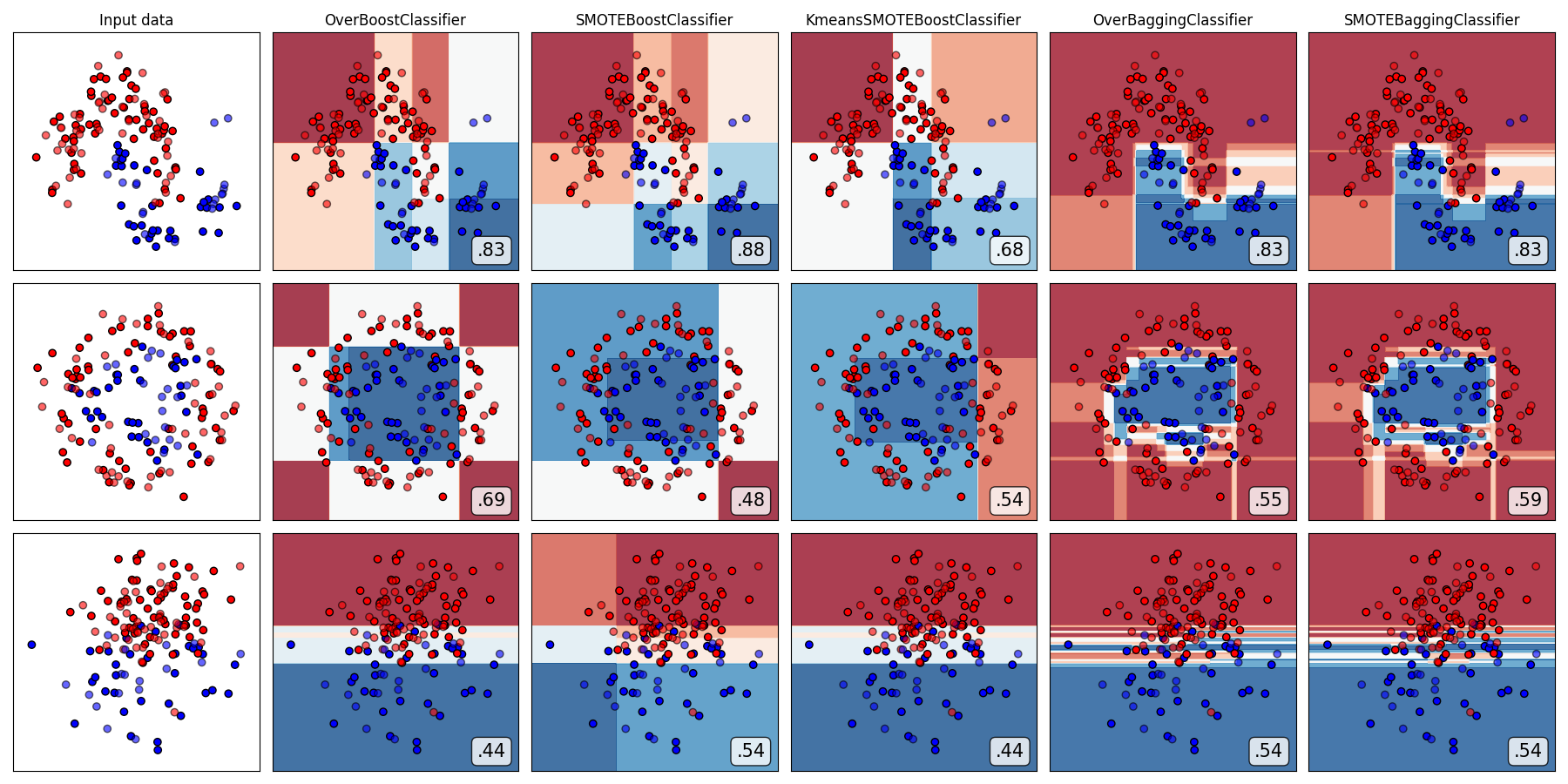
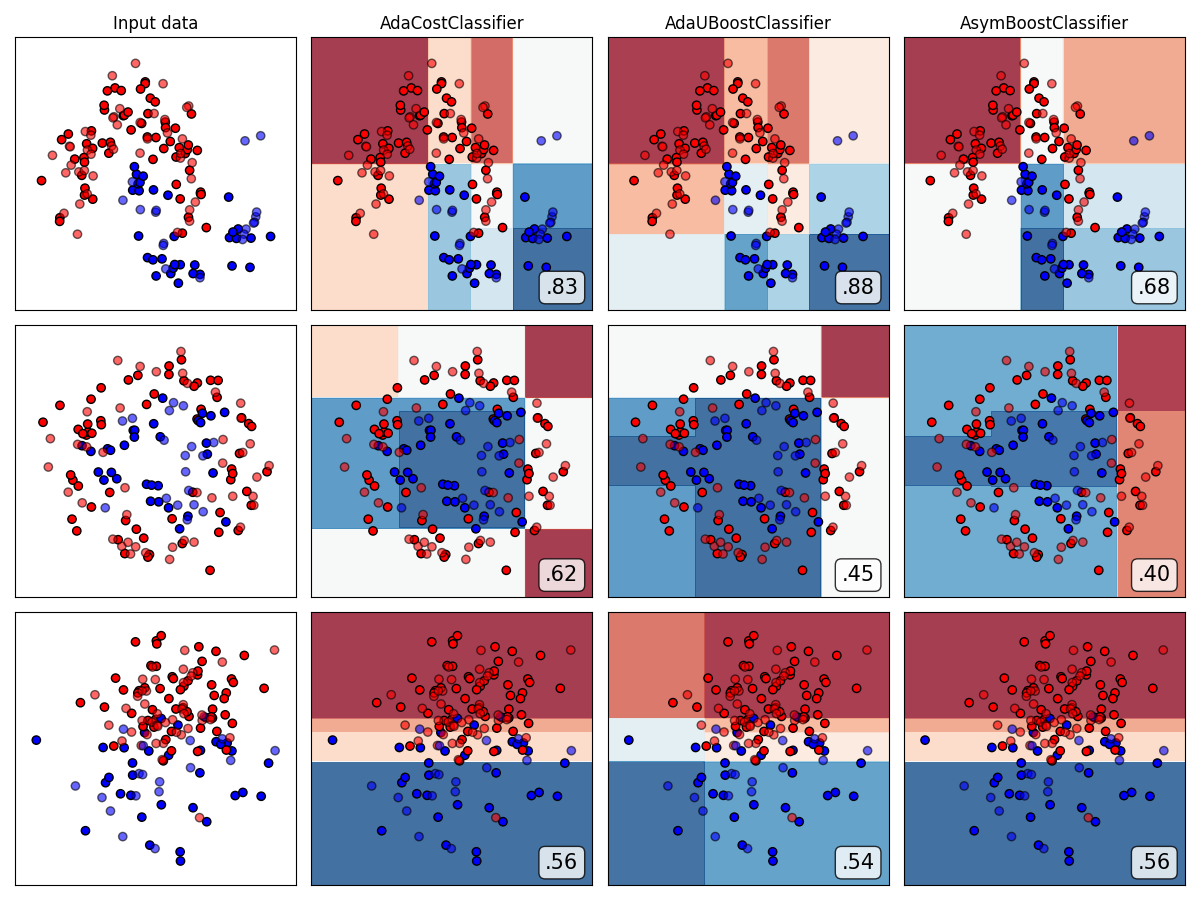
The plots show training points in solid colors and testing points semi-transparent. The lower right shows the average precision score (AUPRC) on the test set.
This example uses:
- Reweighting-based method
# Authors: Zhining Liu <zhining.liu@outlook.com>
# License: MIT
print(__doc__)
# Import imbalanced-ensemble
import imbens
# Import utilities
import numpy as np
import sklearn
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.datasets import make_moons, make_circles, make_classification
from imbens.datasets import make_imbalance
# Import plot utilities
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
RANDOM_STATE = 42
Preparation
Make 3 imbalanced toy classification tasks.
distribution = {0: 100, 1: 50}
# dataset 1
X, y = make_moons(200, noise=0.2, random_state=RANDOM_STATE)
dataset1 = make_imbalance(
X, y, sampling_strategy=distribution, random_state=RANDOM_STATE
)
# dataset 2
X, y = make_circles(200, noise=0.2, factor=0.5, random_state=RANDOM_STATE)
dataset2 = make_imbalance(
X, y, sampling_strategy=distribution, random_state=RANDOM_STATE
)
# dataset 3
X, y = make_classification(
200,
n_features=2,
n_redundant=0,
n_informative=2,
random_state=1,
n_clusters_per_class=1,
)
X += 2 * np.random.RandomState(RANDOM_STATE).uniform(size=X.shape)
dataset3 = make_imbalance(
X, y, sampling_strategy=distribution, random_state=RANDOM_STATE
)
datasets = [dataset1, dataset2, dataset3]
Load all ensemble classifiers
from imbens.utils.testing import all_estimators
init_kwargs = {'n_estimators': 5, 'random_state': RANDOM_STATE}
all_ensembles_clf = {
name: ensemble(**init_kwargs) for (name, ensemble) in all_estimators('ensemble')
}
print('{:<30s} | Class \n{:=<120s}'.format('Method', ''))
for (name, ensemble) in all_estimators('ensemble'):
print('{:<30s} | {}'.format(name, ensemble))
Method | Class
========================================================================================================================
AdaCostClassifier | <class 'imbens.ensemble._reweighting.adacost.AdaCostClassifier'>
AdaUBoostClassifier | <class 'imbens.ensemble._reweighting.adauboost.AdaUBoostClassifier'>
AsymBoostClassifier | <class 'imbens.ensemble._reweighting.asymmetric_boost.AsymBoostClassifier'>
BalanceCascadeClassifier | <class 'imbens.ensemble._under_sampling.balance_cascade.BalanceCascadeClassifier'>
BalancedRandomForestClassifier | <class 'imbens.ensemble._under_sampling.balanced_random_forest.BalancedRandomForestClassifier'>
CompatibleAdaBoostClassifier | <class 'imbens.ensemble._compatible.adaboost_compatible.CompatibleAdaBoostClassifier'>
CompatibleBaggingClassifier | <class 'imbens.ensemble._compatible.bagging_compatible.CompatibleBaggingClassifier'>
EasyEnsembleClassifier | <class 'imbens.ensemble._under_sampling.easy_ensemble.EasyEnsembleClassifier'>
KmeansSMOTEBoostClassifier | <class 'imbens.ensemble._over_sampling.kmeans_smote_boost.KmeansSMOTEBoostClassifier'>
OverBaggingClassifier | <class 'imbens.ensemble._over_sampling.over_bagging.OverBaggingClassifier'>
OverBoostClassifier | <class 'imbens.ensemble._over_sampling.over_boost.OverBoostClassifier'>
RUSBoostClassifier | <class 'imbens.ensemble._under_sampling.rus_boost.RUSBoostClassifier'>
SMOTEBaggingClassifier | <class 'imbens.ensemble._over_sampling.smote_bagging.SMOTEBaggingClassifier'>
SMOTEBoostClassifier | <class 'imbens.ensemble._over_sampling.smote_boost.SMOTEBoostClassifier'>
SelfPacedEnsembleClassifier | <class 'imbens.ensemble._under_sampling.self_paced_ensemble.SelfPacedEnsembleClassifier'>
UnderBaggingClassifier | <class 'imbens.ensemble._under_sampling.under_bagging.UnderBaggingClassifier'>
Function for classifier comparison
def plot_classifier_comparison(classifiers, names, datasets, figsize):
h = 0.02 # step size in the mesh
figure = plt.figure(figsize=figsize)
i = 1
# iterate over datasets
for ds_cnt, ds in enumerate(datasets):
# preprocess dataset, split into training and test part
X, y = ds
X = StandardScaler().fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.4, random_state=42
)
x_min, x_max = X[:, 0].min() - 0.5, X[:, 0].max() + 0.5
y_min, y_max = X[:, 1].min() - 0.5, X[:, 1].max() + 0.5
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
# just plot the dataset first
cm = plt.cm.RdBu
cm_bright = ListedColormap(['#FF0000', '#0000FF'])
ax = plt.subplot(len(datasets), len(classifiers) + 1, i)
if ds_cnt == 0:
ax.set_title("Input data")
# Plot the training points
ax.scatter(
X_train[:, 0], X_train[:, 1], c=y_train, cmap=cm_bright, edgecolors='k'
)
# Plot the testing points
ax.scatter(
X_test[:, 0],
X_test[:, 1],
c=y_test,
cmap=cm_bright,
alpha=0.6,
edgecolors='k',
)
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
ax.set_xticks(())
ax.set_yticks(())
i += 1
# iterate over classifiers
for name, clf in zip(names, classifiers):
ax = plt.subplot(len(datasets), len(classifiers) + 1, i)
clf.fit(X_train, y_train)
score = sklearn.metrics.average_precision_score(y_test, clf.predict(X_test))
# Plot the decision boundary. For that, we will assign a color to each
# point in the mesh [x_min, x_max]x[y_min, y_max].
if hasattr(clf, "decision_function"):
Z = clf.decision_function(np.c_[xx.ravel(), yy.ravel()])
else:
Z = clf.predict_proba(np.c_[xx.ravel(), yy.ravel()])[:, 1]
# Put the result into a color plot
Z = Z.reshape(xx.shape)
ax.contourf(xx, yy, Z, cmap=cm, alpha=0.8)
# Plot the training points
ax.scatter(
X_train[:, 0], X_train[:, 1], c=y_train, cmap=cm_bright, edgecolors='k'
)
# Plot the testing points
ax.scatter(
X_test[:, 0],
X_test[:, 1],
c=y_test,
cmap=cm_bright,
edgecolors='k',
alpha=0.6,
)
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
ax.set_xticks(())
ax.set_yticks(())
if ds_cnt == 0:
ax.set_title(name)
ax.text(
0.95,
0.06,
('%.2f' % score).lstrip('0'),
size=15,
bbox=dict(boxstyle='round', alpha=0.8, facecolor='white'),
transform=ax.transAxes,
horizontalalignment='right',
)
i += 1
plt.tight_layout()
plt.show()
Compare all under-sampling-based ensemble algorithms
from imbens.ensemble._under_sampling.__init__ import __all__ as names
classifiers = [all_ensembles_clf[name] for name in names]
plot_classifier_comparison(
classifiers, names, datasets, figsize=(len(names) * 3 + 3, 9)
)
Compare all over-sampling-based ensemble algorithms
from imbens.ensemble._over_sampling.__init__ import __all__ as names
classifiers = [all_ensembles_clf[name] for name in names]
plot_classifier_comparison(
classifiers, names, datasets, figsize=(len(names) * 3 + 3, 9)
)
Compare all reweighting-based ensemble algorithms
from imbens.ensemble._reweighting.__init__ import __all__ as names
classifiers = [all_ensembles_clf[name] for name in names]
plot_classifier_comparison(
classifiers, names, datasets, figsize=(len(names) * 3 + 3, 9)
)
Total running time of the script: ( 0 minutes 6.800 seconds)